#include <stdio.h>#include <math.h>#include <string.h>#include <stdlib.h>#include "libcpixe.h"#include "stop96.h"#include "compilopt.h"

Go to the source code of this file.
Functions | |
| int | readINPUT (const char *InputFileNm, EXP_PARAM *pexppar, EXTRAINFO *pExtraInfo) |
| int | symbol2Z (char *symbol) |
| int | Z2mass (int Z, double *mass, char option) |
| int | readCOMPOUND (FILE *f, int nelem, COMPOUND *c) |
| int | readsample (char *SampleDefFileNm, int *MaxZ, int *NFoil, foil **Sample) |
| int | createPresentElems (int MaxZ, int NFoil, const foil *MatArray, int **PresentElems) |
| int | readCalcFlags (const char *CalcFlagsFileNm, const int *PresentElems, int MaxZinsample, int AreasFormat, CPIXERESULTS **CalcFlags) |
| int | readEff (const char *CalibFileNm, const int *PresentElems, int MaxZinsample, CalibYld **EffArray) |
| int | readEff2 (const char *CalibFileNm, const int *PresentElems, int MaxZinsample, const CalibYld *LinesEnerArray, CalibYld **EffArray) |
| double | interpolate_efficiency (int ndata, const double *Xarray, const double *Yarray, double X) |
| int | readEff3 (const char *CalibFileNm, const int *PresentElems, int MaxZinsample, const CalibYld *LinesEnerArray, CalibYld **EffArray) |
| int | readLinesEner (const char *LinesEnerFileNm, const int *PresentElems, int MaxZinsample, CalibYld **LinesEnerArray) |
| int | readXYld (const char *XYldFileNm, const int *PresentElems, const CPIXERESULTS *CalcFlags, int MaxZinsample, double *CalEner, XrayYield **XYldArray) |
| int | readFCK (char *FCKCoefFileNm, int MaxZinsample, FluorCKCoef **FCKCoefArray) |
| int | readAbsCoef (const char *TotAbsCoefFileNm, int MaxZinsample, AbsCoef **TotAbsCoefArray) |
| int | readAbsCoef_OLD (char *TotAbsCoefFileNm, int MaxZinsample, const int *PresentElems, const FILTER *Filter, AbsCoef **TotAbsCoefArray) |
| int | createSPTs (const char *path, const EXP_PARAM *pexp, int MaxZinsample, const int *PresentElems, double step, SPT **SPTArray) |
| double | getSP (const COMPOUND *pcmp, const SPT *SPTArray, double E) |
| int | initlyrarray (const EXP_PARAM *pexp, const foil *MatArray, const CalibYld *LinesEnerArray, const AbsCoef *TotAbsCoefArray, const SPT *SPTArray, const int *PresentElems, int NFoil, int *NFoilUsed, LYR **plyrarray) |
| int | initlyr (const EXP_PARAM *pexp, const CalibYld *LinesEnerArray, const AbsCoef *TotAbsCoefArray, const int *PresentElems, LYR *plyr, double *MACoef) |
| int | readFilter (const char *FilterDefFileNm, FILTER *Filter) |
| int | CreateEff (const char *TotAbsCoefFileNm, const char *LinesEnerFileNm, const char *WindowDefFileNm, const char *CrystalDefFileNm, double factor, CalibYld **EffBlockArray, int *dimEffTable, TwoCol **EffTable) |
| int | AllFilterTrans (const char *TotAbsCoefFileNm, const char *LinesEnerFileNm, const char *FilterDefFileNm, CalibYld *AFTArray) |
| int | FilterTrans (int MaxZinsample, const CalibYld *LinesEnerArray, const AbsCoef *TotAbsCoefArray, const int *PresentElems, FILTER *Filter) |
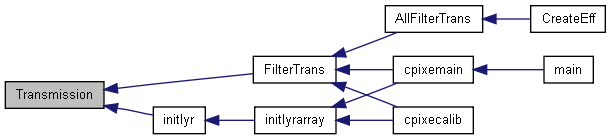
| int | Transmission (const CalibYld *LinesEnerArray, const AbsCoef *TotAbsCoefArray, int Z, const foil *pFoil, double geomcorr, const CalibYld *pTransOld, CalibYld *pTrans) |
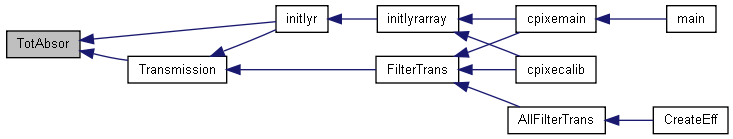
| double | TotAbsor (const AbsCoef *TotAbsCoefArray, const COMPOUND *cmp, double Xray) |
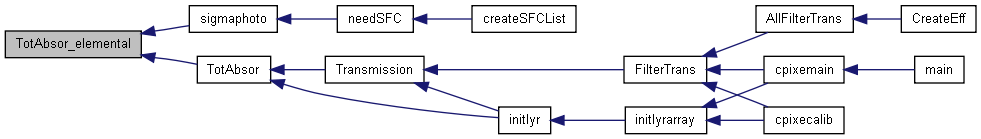
| double | TotAbsor_elemental (int iener, const AbsCoef *Absco, int Z, double Xray) |
| int | createsublyrs (const EXP_PARAM *pexp, const foil *pMat, const SPT *SPTArray, double MajAbsCoef, LYR *plyr) |
| int | SSThick (const EXP_PARAM *pexp, const foil *pMat, const SPT *SPTArray, double MajAbsCoef, double LayerThickness, ESxType ESxin, ESxType *ESxfin, int *pFpos, double *thick, ESxType **ESxA) |
| int | integrate_Simpson2 (const EXP_PARAM *pexp, const AbsCoef *TotAbsCoefArray, const FluorCKCoef *FCKCoefArray, const CalibYld *RawEffiArray, int NFoilUsed, const FILTER *Filter, LYR *plyrarray, CalibYld *XYldSums) |
| int | integrate_Simpson (const EXP_PARAM *pexp, const AbsCoef *TotAbsCoefArray, const FluorCKCoef *FCKCoefArray, const XrayYield *XYldArray, int NFoilUsed, const FILTER *Filter, LYR *plyrarray, CalibYld *XYldSums) |
| int | Xprod (const AbsCoef *AbsC, const FluorCKCoef *pFCK, atomicnumber Z1, atomicnumber Z2, double M2, double ener, CalibYld *XYld) |
| double | PaulX (double ener, atomicnumber z) |
| double | PaulX_y (double MeV, atomicnumber z) |
| double | PaulX_lp (double x, long p) |
| int | ReisX (const AbsCoef *AbsC, const FluorCKCoef *pFCK, double ener, atomicnumber z, double M2, double *sigmaXL) |
| double | ReisX_gs (FwTipo T, int SS, double ksis) |
| double | ReisX_hs (FwTipo T, int SS, double ksih, double thet) |
| double | ReisX_En (double z, int niu) |
| double | ReisX_polisec (int ssind, double kz, double tz) |
| double | ReisX_g (int ss, atomicnumber Zg, double xi) |
| void | PenInteg (atomicnumber atnumb, const CalibYld *AbsFac, const ESxType *ESA, const CalibYld *YldA, const CalibYld *TrsA, const CalibYld *pTrs0, int FExlen, int NeedSFC, int AllowXEqCalc, double x0, double CosInc, CalibYld *XYld, CalibYld *XSFCr, CalibYld *XYldxmed) |
| double | Simps (double a, double b, double fa, double fi, double fb) |
| void | deNormalize (const EXP_PARAM *pexp, const AbsCoef *AbsC, const FluorCKCoef *pFCK, atomicnumber Z2, double M2, double attfraction, const CalibYld *pXYld, XrayYield *ResY, CalibYld *XYldSum) |
| void | deNormalize2 (const EXP_PARAM *pexp, atomicnumber Z2, double attfraction, const CalibYld *pEff, const CalibYld *PenInteg, CalibYld *XYld, CalibYld *XYldSum) |
| void | freeFilter (FILTER *Filter) |
| void | freeExtraInfo (EXTRAINFO *extrainfo) |
| void | freeReusable (int NFoils, LYR **plyrarray, foil **MatArray, CalibYld **XYldSums) |
| void | safefree (void **ptr) |
| void | fprintCALIBYLD (FILE *f, const CalibYld *pCYld) |
| void | fscanCALIBYLD (FILE *f, CalibYld *pCYld) |
| int | compare_Twocol_x (const TwoCol *a, const TwoCol *b) |
News, documentation and latest releases can be found at: http://cpixe.sourceforge.net
A number of papers will be published and should be cited if libcpixe is used for producing scientific publications.
The reference paper for LibCPIXE and CPIXE is: "LibCPIXE: an open-source library for PIXE simulation and Analysis", Carlos Pascual-Izarra, Nuno P. Barradas and Miguel A. Reis, To be published in Proceedings of the IBA2005 (NIMB)
A paper showing the integration of LibCPIXE into the Data Furnace code is: "Simultaneous PIXE and RBS analysis using Bayesian Inference" Carlos Pascual-Izarra, Miguel A. Reis and Nuno P. Barradas, To be published in Proceedings of the IBA2005 (NIMB)
The seed for this code is on a translation from Pascal to C of parts of the code writen by M.A. Reis for the DatPixe-v5.3 code. The Pascal code was initially translated using p2c (an automatic Pascal-to-C free translator) and afterwards heavily modified.
Definition in file libcpixe.c.
| int AllFilterTrans | ( | const char * | TotAbsCoefFileNm, | |
| const char * | LinesEnerFileNm, | |||
| const char * | FilterDefFileNm, | |||
| CalibYld * | AFTArray | |||
| ) |
Returns the transmission factors for all the lines of all the elements (up to maxZ, which is fixed and returned by the function) Note: (Python friendly)
| TotAbsCoefFileNm | (I) Name of the file containing absorption coefs. Coefs are expected in (cm2/mg) | |
| LinesEnerFileNm | (I) Name of the file containing Line energies (in keV). | |
| FilterDefFileNm | (I) Name of the filter definition file. | |
| AFTArray | (O) Pointer to an array of CalibYld structures containing the Transmission factors for each element. |
Definition at line 2378 of file libcpixe.c.
References FilterTrans(), freeFilter(), readAbsCoef(), readFilter(), readLinesEner(), and FILTER::Trans.
Referenced by CreateEff().
Compares TwoCol estructures acording to their x value (used for sorting them)
Definition at line 4311 of file libcpixe.c.
References TwoCol::x.
Referenced by CreateEff().

| int CreateEff | ( | const char * | TotAbsCoefFileNm, | |
| const char * | LinesEnerFileNm, | |||
| const char * | WindowDefFileNm, | |||
| const char * | CrystalDefFileNm, | |||
| double | factor, | |||
| CalibYld ** | EffBlockArray, | |||
| int * | dimEffTable, | |||
| TwoCol ** | EffTable | |||
| ) |
Returns the efficiency of a detector for all the lines in the xener database. The detector is described by two files: one describes the window (inactive layers) and the other the active crystal.
The efficiency is calculated for each given line as Eff=T_w*A_c*factor , where: "T_w" is the transmission of the window for this line, "A_c" is the absorption the active crystal and "factor" is just a scaling factor (if factor=1 , this means it is an ideal 4PI detector)
IMPORTANT: This function is NOT optimized for speed. If speed is needed (e.g. for a fit of the detector characteristics), a new version should be written so that no files are read.
| TotAbsCoefFileNm | (I) Name of the file containing absorption coefs. Coefs are expected in (cm2/mg) | |
| LinesEnerFileNm | (I) Name of the file containing Line energies (in keV) | |
| WindowDefFileNm | (I) Name of a filter definition file describing the detector window | |
| CrystalDefFileNm | (I) Name of a filter definition file describing the detector active area | |
| factor | (I) A scaling factor for the efficiencies (factor=1 means ideal 4PI detector) IMPORTANT!: Use factor=1. (See note 1) | |
| EffBlockArray | (O) Pointer to an array of CalibYld structures containing the Efficiencies factors for each line of each element. | |
| dimEffTable | (O) Dimension of the EffTable array | |
| EffTable | (O) Pointer to an array of TwoCol structures containing Energy (.x) and efficiency (.y) for each line. EffTable is sorted by energy. |
Definition at line 2272 of file libcpixe.c.
References AllFilterTrans(), ChemicalSymbol, compare_Twocol_x(), fprintCALIBYLD(), CalibYld::K_, CalibYld::L_, CalibYld::M_, readLinesEner(), TwoCol::x, and TwoCol::y.
| int createPresentElems | ( | int | MaxZ, | |
| int | NFoil, | |||
| const foil * | MatArray, | |||
| int ** | PresentElems | |||
| ) |
Creates an array of integer flags indicating whether an element is present in the sample (1) or not (0). The dimension of the array is that of the maximum atomic number present in the sample. For example, if the sample contains only C and Si, the array will be: PresentElems[6]= PresentElems[14]=1 (and all the other=0)
| MaxZ | (I) Largest atomic number present in sample (Dimension of PresentElems). | |
| NFoil | (I) Number of target foils | |
| MatArray | (I) Array of foils defining the current sample | |
| PresentElems | (O) Pointer to array of flags indicating presence for each element |
Definition at line 640 of file libcpixe.c.
References foil::comp, COMPOUND::elem, COMPOUND::nelem, and ELEMENT::Z.
Referenced by cpixecalib(), CPIXEMAIN(), cpixemain(), and readFilter().
| int createSPTs | ( | const char * | path, | |
| const EXP_PARAM * | pexp, | |||
| int | MaxZinsample, | |||
| const int * | PresentElems, | |||
| double | step, | |||
| SPT ** | SPTArray | |||
| ) |
Generates the stopping force tables Returns an array of pointers referencing to one table for each element present in sample)
IMPORTANT: The tables are generated in the following units: keV/(1e15at/cm2) !!!
| path | (I) String containing the path to look for the data base files. (e.g. for SCOEF.95A and SCOEF95B in the case of ZBL96 stoppings) Note that the path MUST contain the last "directory separator" | |
| pexp | (I) Pointer to structure containing experimental parameters. | |
| MaxZinsample | (I) Largest atomic number present in sample. | |
| PresentElems | (I) Array of flags indicating presence for each element. See createPresentElems() | |
| step | (I) Energy step to be used (in keV) in the case of linear energy scale or the multiplicative factor in the case logarithmic energy scale. | |
| SPTArray | (O) Array of Stopping power table structs for each present element. IMPORTANT: The tables are generated in the following units: keV/(1e15at/cm2) !!! |
<For the moment, only linear Energy scale is implemented
Definition at line 1921 of file libcpixe.c.
References EXP_PARAM::BeamEner, SPT::dSdE, SPT::E, SPT::Emax, SPT::Estep, FILENMLENGTH, ELEMENT::IM, EXP_PARAM::ion, SPT::logmode, SPT::nrows, nuclearstopping_ZBL(), readstopcoef(), SPT::S, SPT::Smax, stop96d(), and ELEMENT::Z.
Referenced by cpixecalib(), CPIXEMAIN(), and cpixemain().
| int createsublyrs | ( | const EXP_PARAM * | pexp, | |
| const foil * | pMat, | |||
| const SPT * | SPTArray, | |||
| double | MajAbsCoef, | |||
| LYR * | plyr | |||
| ) |
Manages the creation of internal sublayers in each layer
| pexp | (I) Pointer to structure containing experimental parameters. | |
| pMat | (I) Pointer to current foil structure. | |
| SPTArray | (I) Pointer to precalculated tables of stoppings (see createSPT() ) | |
| MajAbsCoef | (I) Maximum absorption coef for this layer. | |
| plyr | (I+O) Pointer to current layer struct. |
Definition at line 2618 of file libcpixe.c.
References foil::comp, EXP_PARAM::cosInc, COMPOUND::elem, ESxType::ep, LYR::ESxArray, LYR::FESxlen, EXP_PARAM::FinalEner, LYR::FoilInEner, LYR::FoilOutEner, getSP(), foil::nfoilelm, SPT::Smax, SSThick(), ESxType::stpp, foil::thick, LYR::ThickIn, ESxType::x, COMPOUND::xn, and ELEMENT::Z.
Referenced by initlyrarray().


| void deNormalize | ( | const EXP_PARAM * | pexp, | |
| const AbsCoef * | AbsC, | |||
| const FluorCKCoef * | pFCK, | |||
| atomicnumber | Z2, | |||
| double | M2, | |||
| double | attfraction, | |||
| const CalibYld * | pXYld, | |||
| XrayYield * | ResY, | |||
| CalibYld * | XYldSum | |||
| ) |
Calculates the yield values using the penetration integral results, i.e. Applies all the factors such as solid angle, atomic fraction, charge,... etc
See eq. (1) of "Reis et al. NIM B 109/110 (1996) 134-138"
Note: This function is only valid for 10 Z2 in range [10,99]
| pexp | (I) Pointer to structure containing experimental parameters. | |
| AbsC | (I) Pointer to Absorption coef structure for element with Z2 | |
| pFCK | (I) pointer to Fluorescence and Coster-Kronig coefficients for Z2 | |
| Z2 | (I) atomic number of the target element | |
| M2 | (I) mass of the target element | |
| attfraction | (I) atomic fraction of the target element in the current layer | |
| pXYld | (I) Calibration Yields for lines corresponding to element Z2 (see note 2) | |
| ResY | (I+O) The calculated yields (in counts) for each layer will be returned in ResY->XYld. Note that ResY->XYld is also an input var (See note 3 below) | |
| XYldSum | (O) The total yield (sum for all layers processed up to this moment of ResY->XYld) is acumulated in XYldSum. |
Note2 The calibration yields are the expected number of counts for a thin sample (of 1e15at/cm2) for 1uC of integrated charge.
Definition at line 3985 of file libcpixe.c.
References XrayYield::atnum, SIM_PARAM::CalEner, SIM_PARAM::ColCharge, EXP_PARAM::DetColFac, SIM_PARAM::DTCC, EXP_PARAM::ion, CalibYld::K_, CalibYld::L_, CalibYld::M_, maxK, minL, minM, EXP_PARAM::simpar, SIM_PARAM::useFilter, Xprod(), XrayYield::XYld, and ELEMENT::Z.
Referenced by integrate_Simpson().

| void deNormalize2 | ( | const EXP_PARAM * | pexp, | |
| atomicnumber | Z2, | |||
| double | attfraction, | |||
| const CalibYld * | pEff, | |||
| const CalibYld * | PenInteg, | |||
| CalibYld * | XYld, | |||
| CalibYld * | XYldSum | |||
| ) |
Calculates the yield values using the penetration integral results, i.e. Applies all the factors such as solid angle, atomic fraction, charge,... etc This function is simmilar to deNormalize() but tries to be less clumsy and more general. It uses the concept of "detector efficiency" instead of "Calibration Yield" (which was an inheritance of the DATTPIXE code).
| pexp | (I) Pointer to structure containing experimental parameters. | |
| Z2 | (I) Atomic number of the target element whose yield is calculated here | |
| attfraction | (I) atomic fraction of the target element in the current layer | |
| pEff | (I) detector efficiencies for the lines corresponding to the target element | |
| PenInteg | (I) Penetration integral results for the lines corresponding to the target element in the current layer | |
| XYld | (O) Calculated Yields (in counts) due to the current layer for the lines corresponding to the target element. | |
| XYldSum | (O) Accumulated Yields (for all layers procesed up to the moment) for the lines corresponding to the target element. |
Yield_ijl=OmegaFraction * Fluence * Eff_j * X_il * PenInteg_ijl
...where the penetration integral (calculated elsewhere) is defined here as: PenInteg_ijl = T_lj * Integral(from E_l,out to E_l,in){T_lj(E) * sigma_ij(E) / S_l(E) dE}
...and where: i-->element j-->line l-->layer OmegaFraction: solid angle fraction(i.e. solid angle of the detector divided by the solid angle of the full sphere (4PI steradians)) Fluence: number of beam particles arriving to the detector DURING the electronics live time (i.e. corrected by the livetime). Can be calculated as Livetime*Collected_charge/Particle_charge. Eff_j: Efficiency of the detector for the j line (taking into account the "intrinsic efficiency, the crystal thickness and the detector window+deadlayer+whatever filtering effect).
Note that the total yield produced by the line j for the element i can be calculated as: totalYield_i,j=SUM_l{Yield_ijl}
Definition at line 4136 of file libcpixe.c.
References EXP_PARAM::Fluence, CalibYld::K_, CalibYld::L_, CalibYld::M_, maxK, minK, minL, minM, EXP_PARAM::SAngFract, and EXP_PARAM::simpar.
Referenced by integrate_Simpson2().

| int FilterTrans | ( | int | MaxZinsample, | |
| const CalibYld * | LinesEnerArray, | |||
| const AbsCoef * | TotAbsCoefArray, | |||
| const int * | PresentElems, | |||
| FILTER * | Filter | |||
| ) |
Calculates the Transmission coefficient of the whole filter for each line that might come out from the "present elements"
| MaxZinsample | (I) Largest atomic number present in sample (dimension of PresentElems). | |
| LinesEnerArray | (I) Array containing the energy of each line. See readXYld() or readEff() | |
| TotAbsCoefArray | (I) Array containing a tables of Absorption Coefs (see readAbsCoef() ) | |
| PresentElems | (I) Array of flags indicating presence for each element. See createPresentElems() | |
| Filter | (I+O) Pointer to filter structure. Output |
Definition at line 2419 of file libcpixe.c.
References FILTER::dimTrans, FILTER::foil, FILTER::geomcorr, FILTER::nlyr, FILTER::Trans, and Transmission().
Referenced by AllFilterTrans(), cpixecalib(), and cpixemain().

| void fprintCALIBYLD | ( | FILE * | f, | |
| const CalibYld * | pCYld | |||
| ) |
Prints a CalibYld structure on a given stream
| f | (I+O) Stream to which the structure is printed | |
| pCYld | (I) Structure to print |
Definition at line 4275 of file libcpixe.c.
References CalibYld::K_, CalibYld::L_, and CalibYld::M_.
Referenced by cpixecalib(), CreateEff(), readCalcFlags(), readEff(), readEff2(), readEff3(), and readLinesEner().
| void freeExtraInfo | ( | EXTRAINFO * | extrainfo | ) |
Frees all memory allocated for the ExtraInfo structure.
| extrainfo | (I+O) Pointer to ExtraInfo structure. |
Definition at line 4202 of file libcpixe.c.
References EXTRAINFO::AreasFileNm, EXTRAINFO::CalibFileNm, EXTRAINFO::DBpath, EXTRAINFO::DetectorFileNm, EXTRAINFO::FilterFileNm, EXTRAINFO::OutputFileNm, safefree(), and EXTRAINFO::SampleFileNm.
Referenced by cpixecalib(), and cpixemain().

| void freeFilter | ( | FILTER * | Filter | ) |
Frees all memory allocated for the filter structure.
| Filter | (I+O) Pointer to filter structure. |
Definition at line 4179 of file libcpixe.c.
References foil::comp, COMPOUND::elem, FILTER::FilterElems, FILTER::foil, FILTER::nlyr, safefree(), FILTER::Trans, COMPOUND::w, COMPOUND::X, and COMPOUND::xn.
Referenced by AllFilterTrans(), cpixecalib(), CPIXEMAIN(), and cpixemain().

Frees the space allocated for arrays that are to be reused in subsequent iterations
| NFoils | (I) Number of target foils | |
| plyrarray | (I+O) Pointer to array of layers. Dim=NFoils+1 | |
| MatArray | (I+O) Pointer to array of foils. Dim=NFoils | |
| XYldSums | (I+O) Pointer to the Output Array of summed X-Ray Yields |
Definition at line 4220 of file libcpixe.c.
References foil::comp, COMPOUND::elem, LYR::ESxArray, LYR::NeedSFC, LYR::ResYldArray, safefree(), LYR::SecResArray, LYR::SSTrsArray, LYR::SSYldArray, LYR::Trans, LYR::TrcAtnum, LYR::TrcUse, COMPOUND::w, COMPOUND::X, and COMPOUND::xn.
Referenced by cpixecalib(), CPIXEMAIN(), and cpixemain().

| void fscanCALIBYLD | ( | FILE * | f, | |
| CalibYld * | pCYld | |||
| ) |
Reads a CalibYld structure from a given stream. The expected format is that as printed by fprintCALIBYLD
| f | (I) Stream from which the structure is read | |
| pCYld | (O) Structure to store the read values |
Definition at line 4294 of file libcpixe.c.
References CalibYld::K_, CalibYld::L_, and CalibYld::M_.
Referenced by readEff(), readEff3(), and readLinesEner().
Returns stopping Using previously calculated tables. Performs a linear interpolation and a Bragg adition.
| pcmp | (I) Pointer to target definition | |
| SPTArray | (I) Pointer to precalculated tables of stoppings (see createSPT() ) | |
| E | (I) Energy of the ion, in keV |
Definition at line 1992 of file libcpixe.c.
References SPT::dSdE, SPT::E, COMPOUND::elem, SPT::Estep, COMPOUND::nelem, SPT::S, COMPOUND::xn, and ELEMENT::Z.
Referenced by createsublyrs(), and SSThick().
| int initlyr | ( | const EXP_PARAM * | pexp, | |
| const CalibYld * | LinesEnerArray, | |||
| const AbsCoef * | TotAbsCoefArray, | |||
| const int * | PresentElems, | |||
| LYR * | plyr, | |||
| double * | MACoef | |||
| ) |
Initializes ONE given layer. It allocates space for various arrays and sets initial values for TrcUse, ResYldArray,... It also calculates the absorption and returns the Maximum absorption coefficient (MACoef).
Note: it uses the global vars minK, maxK, minM, minL,...
| pexp | (I) Pointer to structure containing experimental parameters. | |
| LinesEnerArray | (I) Array containing the energy of each line. See readXYld() or readEff() | |
| TotAbsCoefArray | (I) Array containing a tables of Absorption Coefs (see readAbsCoef() ) | |
| plyr | (I+O) Pointer to current layer struct. | |
| PresentElems | (O) Pointer to array of flags indicating presence for each element | |
| MACoef | (I+O) Maximum absorption coefficient to be used in calcs (presently only cares about K&Lcoefs) |
CAUTION: Change with respect to CDT: TrcUse now starts with index 0
CAUTION: Change with respect to CDT: NeedSFC now starts with index 0
Note:
CAUTION: Change with respect to CDT: TrcAtnum now starts with index 0
Definition at line 2087 of file libcpixe.c.
References SRType::AbsFact, PeakArea::area, PeakArea::atnum, XrayYield::atnum, foil::comp, EXP_PARAM::cosDet, LYR::dimTrans, COMPOUND::elem, XrayYield::ener, CalibYld::K_, CalibYld::L_, CalibYld::M_, maxK, SIM_PARAM::MaxZinsample, minK, minL, minM, LYR::NeedSFC, foil::nfoilelm, LYR::NumOfTrc, LYR::pFoil, SecResType::Pk, LYR::ResYldArray, LYR::SecResArray, EXP_PARAM::simpar, SecResType::SR, TotAbsor(), LYR::Trans, Transmission(), LYR::TrcAtnum, LYR::TrcUse, and ELEMENT::Z.
Referenced by initlyrarray().

| int initlyrarray | ( | const EXP_PARAM * | pexp, | |
| const foil * | MatArray, | |||
| const CalibYld * | LinesEnerArray, | |||
| const AbsCoef * | TotAbsCoefArray, | |||
| const SPT * | SPTArray, | |||
| const int * | PresentElems, | |||
| int | NFoil, | |||
| int * | NFoilUsed, | |||
| LYR ** | plyrarray | |||
| ) |
Initializes the lyr array
| pexp | (I) Pointer to structure containing experimental parameters. | |
| MatArray | (I) Array of foils defining the current sample | |
| LinesEnerArray | (I) Array containing the energy of each line. See readXYld() or readEff() | |
| TotAbsCoefArray | (I) Array containing a tables of Absorption Coefs (see readAbsCoef() ) | |
| SPTArray | (I) Pointer to precalculated tables of stoppings (see createSPT() ) | |
| NFoil | (I) Number of target foils | |
| PresentElems | (O) Pointer to array of flags indicating presence for each element | |
| NFoilUsed | (O) Number of foils finally used in the calcs | |
| plyrarray | (O) Pointer to the layer array. |
Definition at line 2022 of file libcpixe.c.
References LYR::absolutepos, EXP_PARAM::BeamEner, createsublyrs(), EXP_PARAM::FinalEner, LYR::FoilInEner, LYR::FoilOutEner, initlyr(), LYR::pFoil, and LYR::ThickIn.
Referenced by cpixecalib(), and cpixemain().

| int integrate_Simpson | ( | const EXP_PARAM * | pexp, | |
| const AbsCoef * | TotAbsCoefArray, | |||
| const FluorCKCoef * | FCKCoefArray, | |||
| const XrayYield * | XYldArray, | |||
| int | NFoilUsed, | |||
| const FILTER * | Filter, | |||
| LYR * | plyrarray, | |||
| CalibYld * | XYldSums | |||
| ) |
Performs Calculation of Transmission and Yields and then calculates the Penetration Integral. Uses Simpson integration method.
TotAbsCoefArray XYldSums (O)is a matrix which contains, for each element, the total sum of XRayYlds for the whole sample.
Definition at line 2963 of file libcpixe.c.
References SRType::AbsFact, LYR::absolutepos, SIM_PARAM::AllowXEqCalc, foil::comp, EXP_PARAM::cosFac, EXP_PARAM::cosInc, deNormalize(), ESxType::ep, LYR::ESxArray, LYR::FESxlen, EXP_PARAM::ion, CalibYld::K_, CalibYld::L_, CalibYld::M_, maxK, minL, minM, LYR::NeedSFC, LYR::NumOfTrc, PenInteg(), LYR::pFoil, LYR::ResYldArray, LYR::SecResArray, SRType::SFCr, EXP_PARAM::simpar, SecResType::SR, LYR::SSTrsArray, LYR::SSYldArray, LYR::Trans, FILTER::Trans, LYR::TrcAtnum, LYR::TrcUse, ESxType::x, SRType::Xeq, COMPOUND::xn, Xprod(), XrayYield::XYld, ELEMENT::Z, and Z2mass().
Referenced by CPIXEMAIN().
| int integrate_Simpson2 | ( | const EXP_PARAM * | pexp, | |
| const AbsCoef * | TotAbsCoefArray, | |||
| const FluorCKCoef * | FCKCoefArray, | |||
| const CalibYld * | RawEffiArray, | |||
| int | NFoilUsed, | |||
| const FILTER * | Filter, | |||
| LYR * | plyrarray, | |||
| CalibYld * | XYldSums | |||
| ) |
Performs Calculation of Transmission and Yields and then calculates the Penetration Integral. Uses Simpson integration method. It is identical to integrate_Simpson() but it uses efficiencies instead of Calibration yields.
TotAbsCoefArray XYldSums (O)is a matrix which contains, for each element, the total sum of XRayYlds for the whole sample.
Definition at line 2785 of file libcpixe.c.
References SRType::AbsFact, LYR::absolutepos, SIM_PARAM::AllowXEqCalc, foil::comp, EXP_PARAM::cosFac, EXP_PARAM::cosInc, deNormalize2(), ESxType::ep, LYR::ESxArray, LYR::FESxlen, EXP_PARAM::ion, CalibYld::K_, CalibYld::L_, CalibYld::M_, maxK, minL, minM, LYR::NeedSFC, LYR::NumOfTrc, PenInteg(), LYR::pFoil, LYR::ResYldArray, LYR::SecResArray, SRType::SFCr, EXP_PARAM::simpar, SecResType::SR, LYR::SSTrsArray, LYR::SSYldArray, LYR::Trans, FILTER::Trans, LYR::TrcAtnum, LYR::TrcUse, ESxType::x, SRType::Xeq, COMPOUND::xn, Xprod(), XrayYield::XYld, ELEMENT::Z, and Z2mass().
Referenced by cpixecalib(), and cpixemain().

| double interpolate_efficiency | ( | int | ndata, | |
| const double * | Xarray, | |||
| const double * | Yarray, | |||
| double | X | |||
| ) |
returns a efficiency value for a given energy based on a linear interpolation from a Effi VS Ener table
| ndata | Number of data points | |
| Xarray | Abcissas (Energies, in keV, ) | |
| Yarray | Ordinates (Efficiencies) | |
| X | Value to interpolate. |
Definition at line 1054 of file libcpixe.c.
Referenced by readEff2(), and readEff3().
| double PaulX | ( | double | ener, | |
| atomicnumber | z | |||
| ) |
| ener | ||
| z | Atomic number of Target |
Definition at line 3220 of file libcpixe.c.
References PaulX_lp(), and PaulX_y().
Referenced by Xprod().

| double PaulX_lp | ( | double | x, | |
| long | p | |||
| ) |
| x | ||
| p |
Definition at line 3316 of file libcpixe.c.
Referenced by PaulX().
| double PaulX_y | ( | double | MeV, | |
| atomicnumber | z | |||
| ) |
| MeV | Energy of the ion, in MeV | |
| z | Atomic number of Target |
Definition at line 3294 of file libcpixe.c.
Referenced by PaulX().
| void PenInteg | ( | atomicnumber | atnumb, | |
| const CalibYld * | AbsFac, | |||
| const ESxType * | ESA, | |||
| const CalibYld * | YldA, | |||
| const CalibYld * | TrsA, | |||
| const CalibYld * | pTrs0, | |||
| int | FExlen, | |||
| int | NeedSFC, | |||
| int | AllowXEqCalc, | |||
| double | x0, | |||
| double | CosInc, | |||
| CalibYld * | XYld, | |||
| CalibYld * | XSFCr, | |||
| CalibYld * | XYldxmed | |||
| ) |
Calculates a normalized "Penetration Integral" for a given X-ray emission line of a given element. See "Reis et al. NIM B 109/110 (1996) 134-138" See section 1.4 of "Correccoes de Fluorescencia Secundaria em PIXE" Master Thesis by M.A. Reis
It calculates the integral of eq. (4) of the cited paper except that it does not include the normalization constant "sigmaX_ij(Ep)"
| atnumb | (I) Atomic number of the target element | |
| AbsFac | (I) Absorption coefficients structure for the target element | |
| ESA | (I) Pointer to Array of sublayers in the current layer. | |
| YldA | (I) Array of X-ray production cross section for the target element in each sublayer | |
| TrsA | (I) Array of Transmission coefs for the target element in each sublayer | |
| pTrs0 | (I) Array of Transmission factors due to layers over the one we are considering. | |
| FExlen | (I) Number of sublayers in this layer | |
| NeedSFC | (I) Flag indicating whether secondary fluorescence should be calculated (1) or not (0) | |
| AllowXEqCalc | (I) Flag indicating whether "equivalent thickness" should be calculated (1) or not (0) | |
| x0 | (I) absolute in-going path length, in 1e15at/cm2, from sample surface (Only relevant if AllowXEqCalc=1) | |
| CosInc | Cosine of the incident angle (Only relevant if AllowXEqCalc=1) | |
| XYld | (O) Outputs:Penetration integral results (Whatever stored here is just discarded). | |
| XSFCr | (O) Correction factor applied due to secondary fluorescence (Only relevant if NeedSFC=1) | |
| XYldxmed | (O) Equivalent thickness results (Only relevant if AllowXEqCalc=1) |
Definition at line 3772 of file libcpixe.c.
References CYldNul, ESxType::ep, CalibYld::K_, CalibYld::L_, CalibYld::M_, maxK, minL, minM, Simps(), ESxType::stpp, and ESxType::x.
Referenced by integrate_Simpson(), and integrate_Simpson2().
| int readAbsCoef | ( | const char * | TotAbsCoefFileNm, | |
| int | MaxZinsample, | |||
| AbsCoef ** | TotAbsCoefArray | |||
| ) |
Reads from an ascii file the X-ray Absorption coefficients. The File must have the following format: Note: the index of TotAbsCoefArray starts in 1.
----------------- (any header)
MAXZ [maxZ]
DATA (datablocks) -----------------
The datablock for each element has the following structure:
[atnum] [chemsymb]
(23 coefs each one for a given standard energy (see stdtable in TotAbsor() ) [stdcoef[0]]....[stdcoef[22]]
(9 triplets --1 for K, 3 for L and 5 for M-- describing absorp edges for K, L & M (1+3+5 triplets). First member of triplet is the transition ener, 2nd is abs coef for E below E_edge and 3rd is abs coef for E above E_edge ) [enr[1]] [coefenr[0][0]] [coefenr[0][1]] ... [enr[9]] [coefenr[8][0]] [coefenr[8][1]]
-----------------
| TotAbsCoefFileNm | (I) Name of the file to be read. Coefs are expected in (cm2/mg) | |
| MaxZinsample | (I) Largest atomic number present in sample. | |
| TotAbsCoefArray | (O) Pointer to array of structs containing the Absorption Coefs (in cm2/1e15at ) (see readAbsCoef() ) |
Definition at line 1713 of file libcpixe.c.
References AbsCoef::atnum, AbsCoef::coefenr, AbsCoef::coefstd, AbsCoef::enr, LONGSTRINGLENGTH, symbol2Z(), and Z2mass().
Referenced by AllFilterTrans(), cpixecalib(), CPIXEMAIN(), cpixemain(), and main().
| int readAbsCoef_OLD | ( | char * | TotAbsCoefFileNm, | |
| int | MaxZinsample, | |||
| const int * | PresentElems, | |||
| const FILTER * | Filter, | |||
| AbsCoef ** | TotAbsCoefArray | |||
| ) |
Reads from an ascii file the X-ray Absorption coefficients. OLD VERSION (Deprecated for v>1.0) The File must have the following format: Note: the index of TotAbsCoefArray starts in 1.
----------------- (any header)
MAXZ [maxZ]
DATA (datablocks) -----------------
The datablock for each element has the following structure:
[atnum] [chemsymb]
(23 coefs each one for a given standard energy (see stdtable in TotAbsor() ) [stdcoef[0]]....[stdcoef[22]]
(9 triplets --1 for K, 3 for L and 5 for M-- describing absorp edges for K, L & M (1+3+5 triplets). First member of triplet is the transition ener, 2nd is abs coef for E below E_edge and 3rd is abs coef for E above E_edge ) [enr[1]] [coefenr[0][0]] [coefenr[0][1]] ... [enr[9]] [coefenr[8][0]] [coefenr[8][1]]
-----------------
| TotAbsCoefFileNm | (I) Name of the file to be read. Coefs are expected in (cm2/mg) | |
| MaxZinsample | (I) Largest atomic number present in sample. | |
| PresentElems | (I) Array of flags indicating presence for each element. See createPresentElems() | |
| Filter | (I) Pointer to filter structure. | |
| TotAbsCoefArray | (O) Pointer to array of structs containing the Absorption Coefs (in cm2/1e15at ) (see readAbsCoef() ) |
Definition at line 1828 of file libcpixe.c.
References AbsCoef::atnum, AbsCoef::coefenr, AbsCoef::coefstd, AbsCoef::enr, FILTER::FilterElems, LONGSTRINGLENGTH, symbol2Z(), and Z2mass().
| int readCalcFlags | ( | const char * | CalcFlagsFileNm, | |
| const int * | PresentElems, | |||
| int | MaxZinsample, | |||
| int | AreasFormat, | |||
| CPIXERESULTS ** | CalcFlags | |||
| ) |
Reads files containing the Calculation Flags (i.e. which lines should be calculated) or, alternativelly reads areas files from a file in the DATTPIXE Areas format. The behaviour depends on the DTAreasFlag (see below)
The Format of the file is as follows: ----------------- (any header) DATA (datablocks) -----------------
if AreasFormat=1, The datablock for each element has the following structure:
[ChemSymb] [Kalpha1,2] [Kbeta1] [Kbeta2] [Lalpha1,2] [Lbeta2] [Ll] [Lbeta1] [Lgamma1] [Leta] [Lbeta3] [Lbeta4] [Lgamma3] [Malpha1,2] [Mbeta] [Mgamma]
Note: the flags are: 0 if not to be calculated, 1 (or >1) if should be calculated
If AreasFormat=2 the datablock format is: [ChemSymb] [Kalpha1,2] [err] [Kbeta1] [err] [Kbeta2] [err] [Lalpha1,2] [err] [Lbeta2] [err] [Ll] [err] [Lbeta1] [err] [Lgamma1] [err] [Leta] [err] [Lbeta3] [err] [Lbeta4] [err] [Lgamma3] [err] [Malpha1,2] [err] [Mbeta] [err] [Mgamma] [err]
Note: The information for elements not present in the sample is just ignored.
| CalcFlagsFileNm | (I) Name of the input file | |
| PresentElems | (I) Array of flags indicating presence for each element. See createPresentElems() | |
| MaxZinsample | (I) Largest atomic number present in sample (dimension of PresentElems). | |
| AreasFormat | (I) Defines the format of the Areas File. 1 for raw areas format, 2 for DATTPIXE Areas Format | |
| CalcFlags | (O) Array (dimension is allocated to MaxZinsample+1) of structures containing the flags. |
Definition at line 700 of file libcpixe.c.
References CPIXERESULTS::atnum, CPIXERESULTS::err, fprintCALIBYLD(), CalibYld::K_, CalibYld::L_, LONGSTRINGLENGTH, CalibYld::M_, CPIXERESULTS::simareas, and symbol2Z().
Referenced by cpixecalib(), and cpixemain().

| int readCOMPOUND | ( | FILE * | f, | |
| int | nelem, | |||
| COMPOUND * | c | |||
| ) |
Reads a compound of 'nelem' elements from the stream f and fills with this info the the compound c This is a slightly modified version from that of hstoplib.c It fills the normalized atomic and mass concentration. Mass of the elements is obtained by calling Z2mass()
The data from the stream must have the following format (repeated nelem times): [element] [atomic_concentration] ...
| f | (I) Pointer to a stream from which the compound definition is read. | |
| nelem | (I) Number of elements in the compound | |
| c | (O) Pointer to structure defining the compound |
Definition at line 485 of file libcpixe.c.
References ELEMENT::A, COMPOUND::elem, ELEMENT::IM, ELEMENT::M, COMPOUND::name, COMPOUND::nelem, COMPOUND::sumX, ELEMENT::symbol, symbol2Z(), COMPOUND::w, COMPOUND::X, COMPOUND::xn, ELEMENT::Z, and Z2mass().
Referenced by readFilter(), and readsample().
| int readEff | ( | const char * | CalibFileNm, | |
| const int * | PresentElems, | |||
| int | MaxZinsample, | |||
| CalibYld ** | EffArray | |||
| ) |
Reads from an ascii file the detector efficiencies
Note: the index of EffArray starts in 1. The File must have the following format: ----------------- (any header) EFFIFILEVERSION [Versionnumber] <---See Note2 MAXZ [maxZ]
DATA (datablocks) -----------------
Note2: The version number helps in case of future modifications of the effi file format. For the moment, it must be larger than 1
The datablock for each element has the following structure:
[AtNum] [ChemSymb]
{Efficiencies block}
Where the {blocks} are read with fscanCALIBYLD() which means they are like:
[Kalpha1,2] [Kbeta1] [Kbeta2] [Lalpha1,2] [Lbeta2] [Ll] [Lbeta1] [Lgamma1] [Leta] [Lbeta3] [Lbeta4] [Lgamma3] [Malpha1,2] [Mbeta] [Mgamma]
-----------------
| CalibFileNm | (I) Name of the file to be read. | |
| PresentElems | (I) Array of flags indicating presence for each element. See createPresentElems() | |
| MaxZinsample | (I) Largest atomic number present in sample (dimension of PresentElems). | |
| EffArray | (O) Pointer to array of structs containing the efficiencies for each element |
Definition at line 843 of file libcpixe.c.
References EFFIFILEVERSION1, fprintCALIBYLD(), fscanCALIBYLD(), LONGSTRINGLENGTH, and symbol2Z().
| int readEff2 | ( | const char * | CalibFileNm, | |
| const int * | PresentElems, | |||
| int | MaxZinsample, | |||
| const CalibYld * | LinesEnerArray, | |||
| CalibYld ** | EffArray | |||
| ) |
Same as readEff but instead of rading a file made of blocks, it reads a two column file:
Note: the index of EffArray starts in 1. The File must have the following format: ----------------- (any header) EFFIFILEVERSION [Versionnumber] <---See Note2
DATA [Energy] [Efficiency] -----------------
Energy is in keV. Efficiency is a dimensionless number in the range [0,1] (Note that solid angle is not included in efficiency!!!)
Note2: The version number helps in case of future modifications of the effi file format. For the moment, it must be 2
-----------------
| CalibFileNm | (I) Name of the file to be read. | |
| PresentElems | (I) Array of flags indicating presence for each element. See createPresentElems() | |
| MaxZinsample | (I) Largest atomic number present in sample (dimension of PresentElems). | |
| LinesEnerArray | (I) Array containing the energy of each line. See readLinesEner() | |
| EffArray | (O) Pointer to array of structs containing the efficiencies for each element |
Definition at line 952 of file libcpixe.c.
References EFFIFILEVERSION2, fprintCALIBYLD(), interpolate_efficiency(), CalibYld::K_, CalibYld::L_, LONGSTRINGLENGTH, and CalibYld::M_.

| int readEff3 | ( | const char * | CalibFileNm, | |
| const int * | PresentElems, | |||
| int | MaxZinsample, | |||
| const CalibYld * | LinesEnerArray, | |||
| CalibYld ** | EffArray | |||
| ) |
This is a version that accepts input in BOTH the ways supported by readEff and readEff2 (i.e) both two column (aka numeric) and blocks (aka symbolical)
Note: the index of EffArray starts in 1. The File must have the following format: ----------------- (any header) EFFIFILEVERSION [Versionnumber] <---See Note2
DETECTORCURVE [Energy] [Efficiency] ... ...
EXCEPTIONS [AtNum] [ChemSymb]
{Efficiencies block}
Where the {efficiencies blocks} are read with fscanCALIBYLD() which means they are like:
[Kalpha1,2] [Kbeta1] [Kbeta2] [Lalpha1,2] [Lbeta2] [Ll] [Lbeta1] [Lgamma1] [Leta] [Lbeta3] [Lbeta4] [Lgamma3] [Malpha1,2] [Mbeta] [Mgamma]
-----------------
Note2: The version number helps in case of future modifications of the effi file format. For the moment, it must be 3
-----------------
| CalibFileNm | (I) Name of the file to be read. | |
| PresentElems | (I) Array of flags indicating presence for each element. See createPresentElems() | |
| MaxZinsample | (I) Largest atomic number present in sample (dimension of PresentElems). | |
| LinesEnerArray | (I) Array containing the energy of each line. See readLinesEner() | |
| EffArray | (O) Pointer to array of structs containing the efficiencies for each element |
Definition at line 1123 of file libcpixe.c.
References ChemicalSymbol, EFFIFILEVERSION3, fprintCALIBYLD(), fscanCALIBYLD(), interpolate_efficiency(), CalibYld::K_, CalibYld::L_, LONGSTRINGLENGTH, CalibYld::M_, and symbol2Z().
Referenced by cpixecalib(), and cpixemain().
| int readFCK | ( | char * | FCKCoefFileNm, | |
| int | MaxZinsample, | |||
| FluorCKCoef ** | FCKCoefArray | |||
| ) |
Reads from an ascii file the Fluorescence and Coster-Kronig coefficients. Note: the index of FCKCoefArray starts in 1.
----------------- (any header)
MAXZ [maxZ]
DATA (datablocks) -----------------
The datablock for each element has the following structure:
[atnum] [chemsymb]
(Fluorescence Coeficients) [K] [LI] [LII] [LIII] [MI]...
(Line fractions for:) [Kalpha1,2] [Kbeta1] [Kbeta2] [Lalpha1,2] [Lbeta2] [Ll] [Lbeta1] [Lgamma1] [Leta] [Lbeta3] [Lbeta4] [Lgamma3] [Malpha1,2] [Mbeta] [Mgamma]
-----------------
| FCKCoefFileNm | (I) Name of the file to be read. | |
| MaxZinsample | (I) Largest atomic number present in sample. | |
| FCKCoefArray | (O) Pointer to array of structs containing Fluorescence and C-K Coefs |
Definition at line 1611 of file libcpixe.c.
References FluorCKCoef::atnum, FluorCKCoef::ck, FluorCKCoef::k, CalibYld::K_, CalibYld::L_, LONGSTRINGLENGTH, CalibYld::M_, symbol2Z(), and FluorCKCoef::w.
Referenced by cpixecalib(), CPIXEMAIN(), and cpixemain().

| int readFilter | ( | const char * | FilterDefFileNm, | |
| FILTER * | Filter | |||
| ) |
Initializes a Filter structure by reading the filter foil description from a file Note that the Filter created must be freed with freeFilter() if it is not needed anymore
| FilterDefFileNm | (I) Name of a filter definition file | |
| Filter | (O) Filter Structure to be initialized |
<
Definition at line 2189 of file libcpixe.c.
References foil::comp, createPresentElems(), COMPOUND::elem, FILTER::FilterElems, FILTER::foil, FILTER::geomcorr, LONGSTRINGLENGTH, ELEMENT::M, FILTER::MaxZ, foil::nfoilelm, FILTER::nlyr, readCOMPOUND(), foil::thick, and COMPOUND::xn.
Referenced by AllFilterTrans(), cpixecalib(), and cpixemain().

finish implementation of secondary fluorescence
implement forward geometry
| InputFileNm | (I) Name of the input file (no more than LONGSTRINGLENGTH characters) | |
| pexppar | (O) pointer to the experimental parameter structure | |
| pExtraInfo | (O) pointer to structure containing misc data read from input file |
Definition at line 80 of file libcpixe.c.
References ELEMENT::A, SIM_PARAM::AllowSXFCorr, SIM_PARAM::AllowXEqCalc, EXTRAINFO::AreasFileNm, EXTRAINFO::AreasFormat, EXP_PARAM::BeamCol, EXP_PARAM::BeamCross, EXP_PARAM::BeamEner, EXTRAINFO::CalibFileNm, SIM_PARAM::ColCharge, EXP_PARAM::cosDet, EXP_PARAM::cosFac, EXP_PARAM::cosInc, EXTRAINFO::DBpath, DEG2RAD, EXP_PARAM::DetAng, EXP_PARAM::DetColFac, EXTRAINFO::DetectorFileNm, SIM_PARAM::DTCC, ELECTRONCHARGE_IN_UC, EXTRAINFO::FilterFileNm, EXP_PARAM::FinalEner, EXP_PARAM::Fluence, EXTRAINFO::givendetectorfile, ELEMENT::IM, EXP_PARAM::IncAng, EXP_PARAM::ion, EXP_PARAM::ioncharge, LONGSTRINGLENGTH, ELEMENT::M, EXTRAINFO::OutputFileNm, EXTRAINFO::SampleFileNm, EXP_PARAM::SAngFract, EXP_PARAM::simpar, ELEMENT::symbol, symbol2Z(), EXTRAINFO::useefficiencies, EXTRAINFO::WantOutputfile, ELEMENT::Z, and Z2mass().
Referenced by cpixecalib(), and cpixemain().

| int readLinesEner | ( | const char * | LinesEnerFileNm, | |
| const int * | PresentElems, | |||
| int | MaxZinsample, | |||
| CalibYld ** | LinesEnerArray | |||
| ) |
Reads from an ascii file the Line Energies
Note: the index of LinesEnerArray starts in 1. The File must have the following format: ----------------- (any header)
MAXZ [maxZ]
DATA (datablocks) -----------------
Note2: The version number helps in case of future modifications of the effi file format. For the moment, it must be larger than 1
The datablock for each element has the following structure:
[AtNum] [ChemSymb]
{ELine Energies}
Where the {blocks} are read with fscanCALIBYLD() which means they are like:
[Kalpha1,2] [Kbeta1] [Kbeta2] [Lalpha1,2] [Lbeta2] [Ll] [Lbeta1] [Lgamma1] [Leta] [Lbeta3] [Lbeta4] [Lgamma3] [Malpha1,2] [Mbeta] [Mgamma]
-----------------
| LinesEnerFileNm | (I) Name of the file to be read. | |
| PresentElems | (I) Array of flags indicating presence for each element. See createPresentElems() | |
| MaxZinsample | (I) Largest atomic number present in sample (dimension of PresentElems). | |
| LinesEnerArray | (O) Pointer to array of structs containing the X-ray emmission energies for each element |
Definition at line 1339 of file libcpixe.c.
References fprintCALIBYLD(), fscanCALIBYLD(), LONGSTRINGLENGTH, and symbol2Z().
Referenced by AllFilterTrans(), cpixecalib(), cpixemain(), and CreateEff().
| int readsample | ( | char * | SampleDefFileNm, | |
| int * | MaxZ, | |||
| int * | NFoil, | |||
| foil ** | Sample | |||
| ) |
Allocates and initializes the sample definition from a file. It also returns Number of foils and Maximum atomic number present in sample.
The File must have the following format: ----------------- NUMBER_OF_FOILS [nfoils] FOIL [thickness_in_ug/cm2] [numberofelems] [symbol] [concentration] [symbol] [concentration] ... FOIL CalEner [numberofelems] [symbol] [concentration] ... -----------------
Note:Any number of foils and any number of elements are allowed.
| SampleDefFileNm | (I) Name of the file to be read. | |
| MaxZ | (O) Largest atomic number present in sample (Dimension of PresentElems). | |
| NFoil | (O) Number of target foils | |
| Sample | (O) Pointer to Array of files (Dim=NFoil) |
Definition at line 575 of file libcpixe.c.
References foil::comp, COMPOUND::elem, LONGSTRINGLENGTH, ELEMENT::M, foil::nfoilelm, readCOMPOUND(), foil::thick, and COMPOUND::xn.
Referenced by cpixecalib(), and cpixemain().

| int readXYld | ( | const char * | XYldFileNm, | |
| const int * | PresentElems, | |||
| const CPIXERESULTS * | CalcFlags, | |||
| int | MaxZinsample, | |||
| double * | CalEner, | |||
| XrayYield ** | XYldArray | |||
| ) |
Reads from an ascii file the calibration yields (aka "thin film yields at CalEner") and also returns CalEner.
Note: the index of XYldArray starts in 1. The File must have the following format: ----------------- (any header) CALENER [calener] MAXZ [maxZ]
DATA (datablocks) -----------------
The datablock for each element has the following structure:
[AtNum] [ChemSymb]
[Kalpha1,2] [Kbeta1] [Kbeta2] <----Energies [Kalpha1,2] [Kbeta1] [Kbeta2] <----Yields
[Lalpha1,2] [Lbeta2] [Ll] <---------Energies [Lalpha1,2] [Lbeta2] [Ll] <---------Yields [Lbeta1] [Lgamma1] [Leta] <--------Energies [Lbeta1] [Lgamma1] [Leta] <--------Yields [Lbeta3] [Lbeta4] [Lgamma3] <----Energies [Lbeta3] [Lbeta4] [Lgamma3] <----Yields
[Malpha1,2] [Mbeta] [Mgamma] <-----Energies [Malpha1,2] [Mbeta] [Mgamma] <-----Yields
-----------------
| XYldFileNm | (I) Name of the file to be read. | |
| PresentElems | (I) Array of flags indicating presence for each element. See createPresentElems() | |
| CalcFlags | (I) array of structures indicating which lines are to be calculated (simareas values>0) and which not (simareas values=-1). See readCalcFlags() | |
| MaxZinsample | (I) Largest atomic number present in sample (dimension of PresentElems). | |
| CalEner | (O) Energy at which the calibration was performed | |
| XYldArray | (O) Pointer to array of structs containing the "thin film yields" for each element |
Definition at line 1451 of file libcpixe.c.
References CPIXERESULTS::atnum, XrayYield::atnum, XrayYield::ener, CalibYld::K_, CalibYld::L_, LONGSTRINGLENGTH, CalibYld::M_, CPIXERESULTS::simareas, XrayYield::symb, symbol2Z(), XrayYield::XYld, and Z2mass().
Referenced by CPIXEMAIN().

| int ReisX | ( | const AbsCoef * | AbsC, | |
| const FluorCKCoef * | pFCK, | |||
| double | ener, | |||
| atomicnumber | z, | |||
| double | M2, | |||
| double * | sigmaXL | |||
| ) |
Returns X-ray fluorescence cross-sections for L lines. See paper ???
| AbsC | (I) Pointer to Absorption coef structure for element with Z2 | |
| pFCK | (I) Pointer to structure containing the Fluorescence and Coster-Kronig coefs | |
| ener | (I) Proton energy, in keV | |
| z | (I) Atomic number of Target | |
| M2 | (I) Atomic mass of Target (in amu) | |
| sigmaXL | (O) X-ray cross sections, in barn |
U.Atomicas de massa
U.Atomicas
parametro de correccao de polarizacao-ligacao
correccao de defleccao Coulombiana
Definition at line 3360 of file libcpixe.c.
References BARNTOM2, FluorCKCoef::ck, AbsCoef::enr, L, MEVAMU, ReisX_En(), ReisX_g(), ReisX_gs(), ReisX_hs(), ReisX_polisec(), UATMASS, and FluorCKCoef::w.
Referenced by Xprod().
| double ReisX_En | ( | double | z, | |
| int | niu | |||
| ) |
Auxiliar function to ReisX()
| z | ||
| niu |
aproximation for niu>>
vide Abramowitz & Stegun , Dover 1Ed. 1965
Definition at line 3588 of file libcpixe.c.
Referenced by ReisX().
| double ReisX_g | ( | int | ss, | |
| atomicnumber | Zg, | |||
| double | xi | |||
| ) |
| double ReisX_gs | ( | FwTipo | T, | |
| int | SS, | |||
| double | ksis | |||
| ) |
Auxiliar function to ReisX()
| T | ||
| SS | ||
| ksis |
ref. Brandt and Lapicki,Phys.Rev.,A20(1979)465
Definition at line 3463 of file libcpixe.c.
Referenced by ReisX().
| double ReisX_hs | ( | FwTipo | T, | |
| int | SS, | |||
| double | ksih, | |||
| double | thet | |||
| ) |
| double ReisX_polisec | ( | int | ssind, | |
| double | kz, | |||
| double | tz | |||
| ) |
| void safefree | ( | void ** | ptr | ) |
Frees memory only if *ptr is not NULL
| ptr | (I+O) Pointer to array to be freed |
Definition at line 4260 of file libcpixe.c.
Referenced by CPIXEMAIN(), freeExtraInfo(), freeFilter(), and freeReusable().
| double Simps | ( | double | a, | |
| double | b, | |||
| double | fa, | |||
| double | fi, | |||
| double | fb | |||
| ) |
Returns area of trapezoid, useful for Simpson Integration.
See "Corrected Trapezoidal Rule" in Conte & de Boor "Elementar Numerical Analysis" 3rd Ed. McGraw-Hill,1980,,p309 ExpCTR:=(b-a)/2*(fa+fb)
| a | First point of the trapezoid base | |
| b | Second point of the Trapezoid base | |
| fa | ||
| fi | ||
| fb |
Definition at line 3954 of file libcpixe.c.
Referenced by PenInteg().

| int SSThick | ( | const EXP_PARAM * | pexp, | |
| const foil * | pMat, | |||
| const SPT * | SPTArray, | |||
| double | MajAbsCoef, | |||
| double | LayerThickness, | |||
| ESxType | ESxin, | |||
| ESxType * | ESxfin, | |||
| int * | pFpos, | |||
| double * | thick, | |||
| ESxType ** | ESxA | |||
| ) |
Recursive function which divides a sublayer in two if either energy loss or absorption are too large
| pexp | (I) Pointer to structure containing experimental parameters. | |
| pMat | (I) Pointer to current foil structure. | |
| SPTArray | (I) Pointer to precalculated tables of stoppings (see createSPT() ) | |
| MajAbsCoef | (I) Maximum absorption coef for this layer. | |
| LayerThickness | (I) Effective Thickness (in-going path) for the layer containing this sublayer. | |
| ESxin | (I) Struct of energy, stopping and depth (ESx) for first half of this sublayer | |
| ESxfin | (I+O) Pointer to struct of energy, stopping and depth (ESx) for 2nd half of this sublayer | |
| pFpos | (I+O) Pointer to index of sublayer position into the layer. | |
| thick | (O) Pointer to thickness of current sublayer. | |
| ESxA | (O) Pointer to Array of sublayers in the current layer. |
Definition at line 2695 of file libcpixe.c.
References EXP_PARAM::BeamEner, foil::comp, EXP_PARAM::cosFac, ESxType::ep, getSP(), SSThick(), ESxType::stpp, and ESxType::x.
Referenced by createsublyrs(), and SSThick().

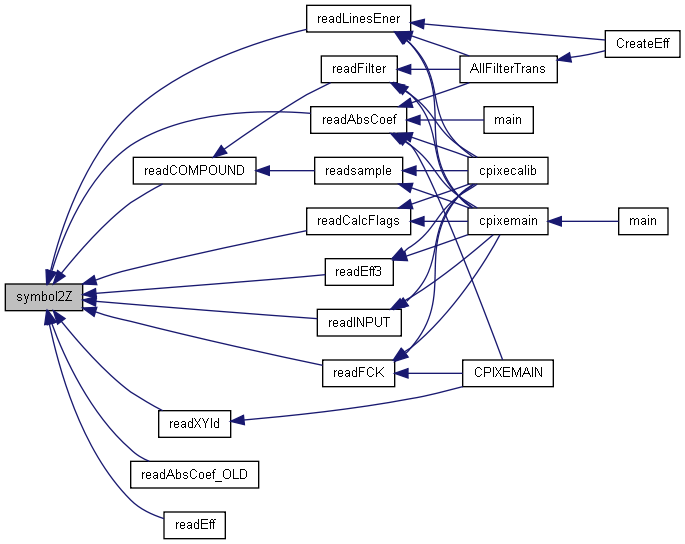
| int symbol2Z | ( | char * | symbol | ) |
Returns the atomic number for a certain element (up to Z=109) This is a faster version than that of hstoplib.c because it does not access any file. It uses the ChemicalSymbol global constant array defined in libcpixe.h
| symbol | (I) Chemical symbol of the element (case insensitive) |
Array containing the chemical symbols from H(Z=1) to Mt(Z=109)
Definition at line 378 of file libcpixe.c.
References ChemicalSymbol.
Referenced by readAbsCoef(), readAbsCoef_OLD(), readCalcFlags(), readCOMPOUND(), readEff(), readEff3(), readFCK(), readINPUT(), readLinesEner(), and readXYld().
Calculates the total absoption (it returns it as a double) for a given X-ray line in a given compound
| TotAbsCoefArray | (I) Array containing a tables of Absorption Coefs (see readAbsCoef() ) | |
| cmp | (I) Definition of the absorber material. | |
| Xray | (I) Energy of the X-ray, in keV |
<
Definition at line 2509 of file libcpixe.c.
References COMPOUND::elem, COMPOUND::nelem, TotAbsor_elemental(), COMPOUND::xn, and ELEMENT::Z.
Referenced by initlyr(), and Transmission().

| double TotAbsor_elemental | ( | int | iener, | |
| const AbsCoef * | Absco, | |||
| int | Z, | |||
| double | Xray | |||
| ) |
Calculates the total absoption coefficient (it returns it as a double) for a given X-ray line in a given element.
| iener | (I) Index of the absorption line to be considered | |
| Absco | (I) Pointer to structure of Absorption Coefs for the absorber element | |
| Z | (I) Atomic number of the absorber element | |
| Xray | (I) Energy of the X-ray, in keV |
Definition at line 2553 of file libcpixe.c.
References AbsCoef::coefenr, AbsCoef::coefstd, and AbsCoef::enr.
Referenced by sigmaphoto(), and TotAbsor().
| int Transmission | ( | const CalibYld * | LinesEnerArray, | |
| const AbsCoef * | TotAbsCoefArray, | |||
| int | Z, | |||
| const foil * | pFoil, | |||
| double | geomcorr, | |||
| const CalibYld * | pTransOld, | |||
| CalibYld * | pTrans | |||
| ) |
Calculates the Transmission coefficients for a given foil (whith effective thickness calculated using a geometric correction).
| LinesEnerArray | (I) Array containing the energy of each line. See readXYld() or readEff() | |
| TotAbsCoefArray | (I) Array containing a tables of Absorption Coefs (see readAbsCoef() ) | |
| Z | (I) Atomic number of the element for whose lines the transmission is calculated | |
| pFoil | (I) pointer to the current foil definition | |
| geomcorr | (I) geometric correction to the foil thickness (allows tiltings) | |
| pTransOld | (I) If this pointer is not null, pTrans will be multiplied by it line-by-line. | |
| pTrans | (O) Structure where the transmission coefs for each line are returned. |
Definition at line 2466 of file libcpixe.c.
References foil::comp, CalibYld::K_, CalibYld::L_, CalibYld::M_, maxK, minL, minM, foil::thick, and TotAbsor().
Referenced by FilterTrans(), and initlyr().

| int Xprod | ( | const AbsCoef * | AbsC, | |
| const FluorCKCoef * | pFCK, | |||
| atomicnumber | Z1, | |||
| atomicnumber | Z2, | |||
| double | M2, | |||
| double | ener, | |||
| CalibYld * | XYld | |||
| ) |
Calculates X-Ray production efficiency for a given beam and target, in cm2/(1e15at).
| AbsC | (I) pointer to Absorption coef structure for Z2 | |
| pFCK | (I) pointer to Fluorescence and Coster-Kronig coefficients for Z2 | |
| Z1 | (I) atomic number of the ion (note: currently only Z1=1 is supported) | |
| Z2 | (I) atomic number of the target | |
| M2 | (I) mass of the target atom | |
| ener | (I) energy of the incident ion, in keV | |
| XYld | (O) Pointer to return the X-ray production efficiency, in cm2/(1e15at) |
Definition at line 3140 of file libcpixe.c.
References CYldNul, FluorCKCoef::k, CalibYld::K_, CalibYld::L_, maxK, minL, minM, PaulX(), and ReisX().
Referenced by deNormalize(), integrate_Simpson(), and integrate_Simpson2().
| int Z2mass | ( | int | Z, | |
| double * | mass, | |||
| char | option | |||
| ) |
Returns mass information for a given element (specified by its atomic number). Currently the range is limited from Z=1 (H) to Z=92 (U). It provides the Natural (average) weight, Most Abundant Isotope (MAI) weight and Most Abundant Isotope number of nucleons
| Z | (I) Atomic number | |
| mass | (O) mass in amu. See also the "option" argument. | |
| option | (I) Either 'n' or 'm' to select "Natural" or "MAI" mass, respectively |
Definition at line 409 of file libcpixe.c.
Referenced by integrate_Simpson(), integrate_Simpson2(), readAbsCoef(), readAbsCoef_OLD(), readCOMPOUND(), readINPUT(), readXYld(), translatesample(), and translatesample2().
 1.5.4
1.5.4